Population genomics
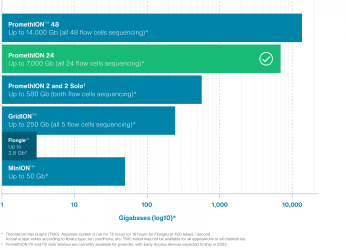
The advent of high-throughput genomic analysis technologies has driven the field of population genomics — the large-scale comparison of genomes within a population. Population genomics has been fundamental to the identification of novel variation that can define specific phenotypes, such as disease susceptibility; however, the technical limitations of traditional analysis platforms may miss important sources of genomic variation. Combining long sequencing reads (up to 4 Mb) with high sample throughput (up to 14 Tb on PromethION*) nanopore technology makes it possible to generate highly contiguous genomes, with resolution and phasing of single nucleotide variants (SNVs), structural variants (SVs), repeats, and DNA methylation in a single assay — across 10’s to 1,000’s of samples.
Long read sequencing of 3,622 Icelanders provides insight into the role of structural variants in human diseases and other traits
Read the publication
We identified over 22,636 SVs per individual, three to five times more than found with short-read sequencing dataBeyter et al. bioRxiv 848366 (2021)